Blogger: Bendik Lund 
During treatment of childhood blood cancer, great variations in side-effects are seen – both in terms of prevalence and seriousness. Some children get more serious side-effects than others. Potentially, the diversity in the toxicity burden for individual patients could reflect the normal genetic variation between patients.
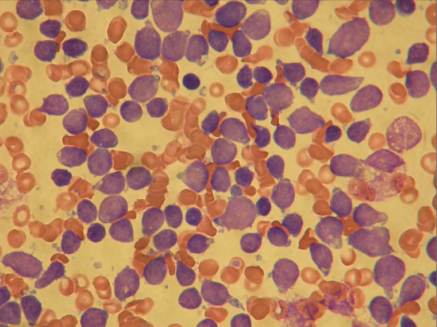
A bone marrow smear at high magnification taken at diagnosis. Most of the blue cells are leukaemic cells. Normal red blood cells are also seen. (Photo: Bendik Lund)
In parallel with the biotechnological development over the last 10-15 years, we have gained extensive knowledge about the normal sequence variation in DNA, which differs from person to person. This sequence variation might explain some of the differences between people, for example height, hair colour, risk of diseases and the body’s reactions to medicines (pharmacogenetics).
There are many types of DNA-variations and one of the most common ones is single nucleotide polymorphism (SNP), where one letter in our genetic code has been replaced by another letter. DNA consists of long chains of base pairs (letters, totalling around 3 billion) and a SNP occurs approximately for every 300th base pair.
We wanted to study what role the natural genetic variation plays in the development of side effects in children treated for leukaemia (cancer of the blood). The most common form of blood cancer in children is acute lymphoblastic leukaemia, and 30-40 children are diagnosed in Norway every year with this type of leukaemia. The treatment consists of chemotherapy given over a period of 2.5 years, and the survival rate today is around 85%. The treatment causes many side effects including reduced immune function and infections. In some cases, the treatment can lead to so serious side effects that the patient dies from the toxicity.
Knowledge about pharmacogenetic variation is already used in the standard treatment for acute lymphoblastic leukaemia when using the chemotherapy 6-mercaptopurine. This drug is dosed based on the patient’s SNP variants for the enzyme that metabolises 6-mercaptopurine (TPMT-genetic variants).
We have collaborated with a research group at the laboratory in Copenhagen (Bonkolab, Rigshospitalet) and, based on existing literature, around 2300 candidate genes that could be significant for children with acute lymphoblastic leukaemia have been identified. Furthermore, the group has made a cost-efficient analysis method where 34,000 genetic variants (SNPs) per patient within these genes (extended candidate gene model) are analysed. Samples from several patients can also be analysed in the same sample tube (multiplexing).
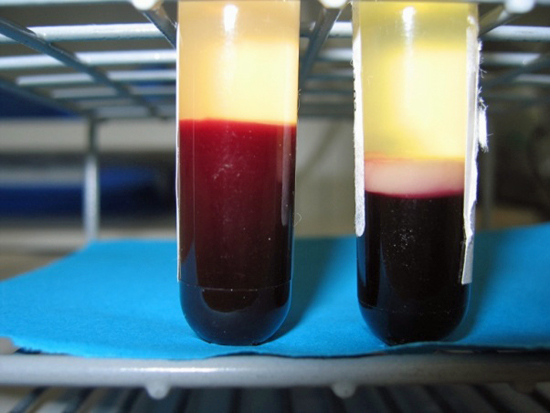
The test tube to the left contains a blood sample from a healthy person. The test tube to the right contains a blood sample form a child with leukaemia. “Leukaemia” means “white blood”, and one can clearly see why when looking at the white layer of cells in the test tube to the right. (Photo: Bendik Lund)
We used this method in a study where we included 69 Danish children with leukaemia and compared the gene variant pattern with clinical data for infections that occurred during the first 50 days of treatment. We identified a SNP profile which with great accuracy can predict the risk for infections in this early phase of the treatment, where many infections are life-threatening.
If these findings are confirmed in similar studies, we may in the future be able to quickly determine whether a patient has an increased risk for serious infections by taking a simple blood test. If the patient is at high risk for serious infections, the treatment could be adapted accordingly for example giving prophylactic antibiotics, or by reducing the intensity of the chemotherapy. Hopefully this will lead to less side effects and higher survival rates.
Further reading:
- Host genome variations and risk of infections during induction treatment for childhood acute lymphoblastic leukaemia
- Risk factors for treatment related mortality in childhood acute lymphoblastic leukaemia
