 By Brooke Wolford, a PhD Candidate from the University of Michigan. Currently working with the K.G. Jebsen Center for Genetic Epidemiology at NTNU
By Brooke Wolford, a PhD Candidate from the University of Michigan. Currently working with the K.G. Jebsen Center for Genetic Epidemiology at NTNU
If you’re from Trøndelag county you probably know someone who has participated in the Nord-Trøndelag Health study (HUNT). Maybe you even participated! But did you know that researchers at the K.G. Jebsen Center for Genetic Epidemiology collaborate with international scientists to study the genetics of diseases like heart disease and diabetes using HUNT?
Genotyping allows researchers to read the DNA nucleotide (e.g. A, T, C, or G) at millions of places in the human genome, and from 2014-2015, DNA from a subset of blood samples donated by HUNT participants was genotyped in an international effort between NTNU’s Dr. Kristian Hveem and Dr. Cristen Willer of the University of Michigan.
Variation in the human genome
The human genome is 3.1 billion nucleotides in size, and these nucleotides ultimately create the proteins that run the human body. Variation in the human genome is normal — I may have a T in a place that you have a C. But sometimes that variation puts a person at increased risk for developing a given disease. Usually this is because a person has inherited a unique combination of risk increasing alleles that in aggregate contribute to disease predisposition.
Genome Wide Association Study (GWAS)
To identify the genetic variants which increase disease risk, scientists use a statistical method called Genome Wide Association Study (GWAS) to identify which nucleotides at which genomic locations are associated with a given disease. Ultimately, identifying and understanding these associations may lead to treatments and pharmaceutical therapies. It also helps clinicians identify people with a high genetic risk score, which means they have a greater genetic burden of risk increasing variants than the average person. It is a current topic of research if people with higher genetic risk scores might benefit from lifestyle interventions and preventative care.
Phenome Wide Association Study (PheWAS)
Scientists from NTNU and the University of Michigan’s Center for Statistical Genetics use HUNT to develop statistical methods and discover novel disease-causing genetic variants. One example of their work is performing GWAS on hundreds of diseases, or phenotypes, in a phenome wide association study (PheWAS). A phenotype is the expressed characteristic in an organism that results from the genotype and the environment. When we consider many or all the phenotypes of an organism, we call this the phenome. PheWAS can identify genomic locations where a nucleotide leads to increased risk for multiple diseases.
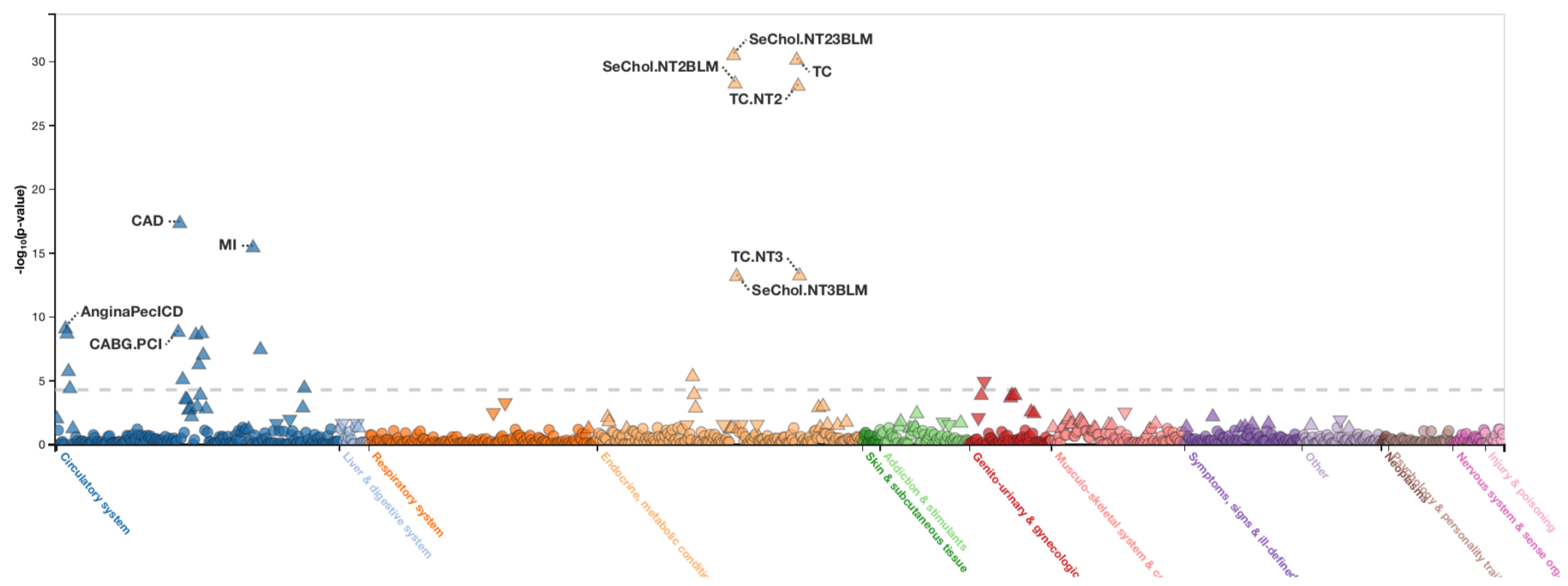
PheWAS was performed on hundreds of diseases like coronary artery disease and laboratory measurements like blood lipid levels across multiple instances of HUNT enrollment. The LPA variant on chromosome 6, rs10455872, is associated with many of these phenotypes—coronary artery disease (CAD), myocardial infarction or heart attack (MI), coronary artery bypass graft which is a type of heart surgery (CABG.PCA), angina or chest pain (AnginaPecICD), and increased blood levels of serum cholesterol (SeChol.NT2BLM, SeChol.NT23BLM, SeChol.NT3BLM), and total cholesterol (TC, TC.NT2, TC.NT3).
For example, a variant in the LPA gene, which codes for the Lp(a) lipoprotein responsible for transporting fats in the body, is known to be associated with heart disease. By performing PheWAS for this LPA variant in HUNT (Figure 1), researchers find the G nucleotide is associated with an increased risk of coronary artery disease, myocardial infarction or heart attack, coronary artery bypass graft which is a type of heart surgery, angina or chest pain, and increased blood levels of serum cholesterol and total cholesterol.
HUNT Data can help us understand diseases
While these associations have been previously noted by clinicians and researchers, PheWAS enables many cross-phenotype comparisons in HUNT. This is important information for understanding disease mechanism and creating potential therapies. PheWAS may also identify a nucleotide that is protective for one disease but increases the risk for another. Because of Norway’s national registries and HUNT’s detailed questionnaires which span decades, HUNT is a valuable resource for international researchers who work to understand how variation in the human genome affects the entire phenome.
