Blogger: Richard Kumaran Kandasamy Associate Professor and Onsager Fellow at Centre of Molecular Inflammation Research (SFF-CEMIR)
Our innate immune system is the first and most important barrier of microbial threats such as viruses and bacteria. It will sense, and in most cases, clear out these pathogens – but not always. A new approach to studying macrophage response to viral threats have resulted in a vastly expanded knowledgebase of the dynamics of the host response to viral infection, and in turn how antiviral innate immunity works. The data is freely available at www.infectome-map.org.
Richard Kumaran Kandasamy.
Photo: Thor Nielsen.
Digging deeper with big data
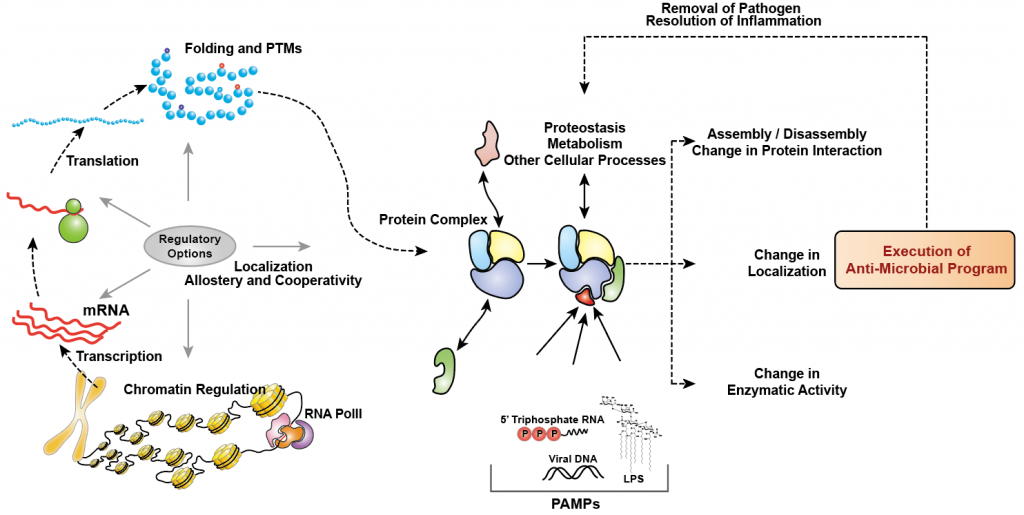
Our immune system is comprised of different types of cells such as macrophages that carry out these specialized tasks of handling the intruder. Although antiviral innate immune response has been widely studied over the past decades and used for development of therapeutics, most of these are based on candidate approach due to the lack of sensitive high-throughput technologies. With the emergence of systems biology and developments in the OMICS technologies (transcriptomics, proteomics and phosphoproteomics etc), the classical view of one-gene-does-everything-in-a-cell is challenged and it is becoming evident that cellular systems are more like a highly connected network that work in a coherent fashion. There are several studies in the recent past that have highlighted that cells indeed have multiple regulatory options (chromatin remodeling, transcription, translation, post-translational modifications (PTMs), folding, cellular localization, etc.) in how it achieves homeostasis under various perturbation scenarios such as viral or bacterial infection (Figure 1).
Using state-of-the-art orthogonal OMICS approaches, we envisioned to understand the dynamics of the host response to viral infection by which we could assess the extent and the molecular logic of the host cellular response. This has the potential to provide unique and complementary information that can allow us to precisely map the systems-level perturbation caused by the viral infection and the viral circumvention of the host response, which will further add to the growing knowledgebase of antiviral innate immunity.
Answers hiding in the shadows of existing research
During our study we learned that post-translational modifications such as phosphorylation are crucial for innate immune response, but also highly understudied.
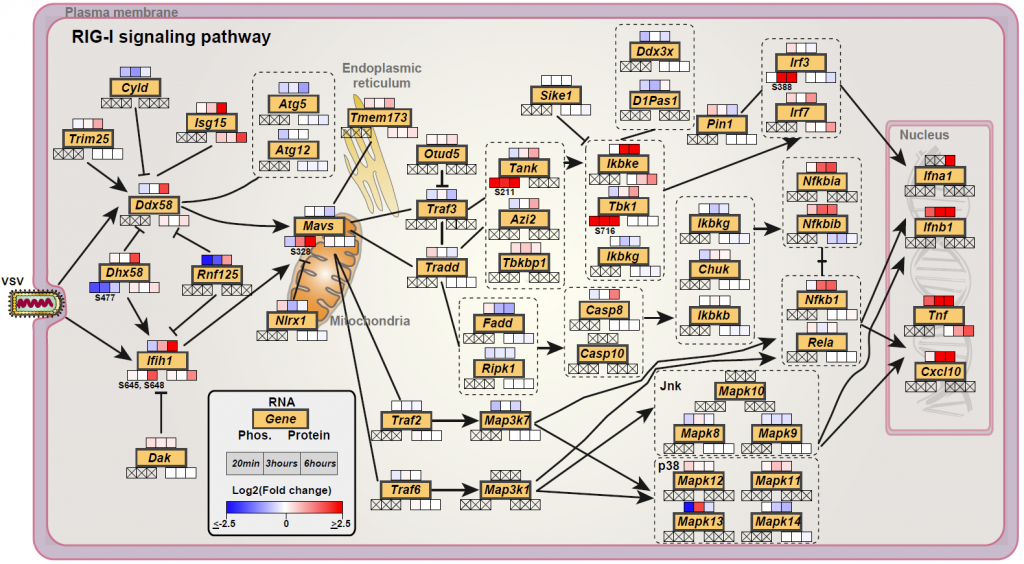
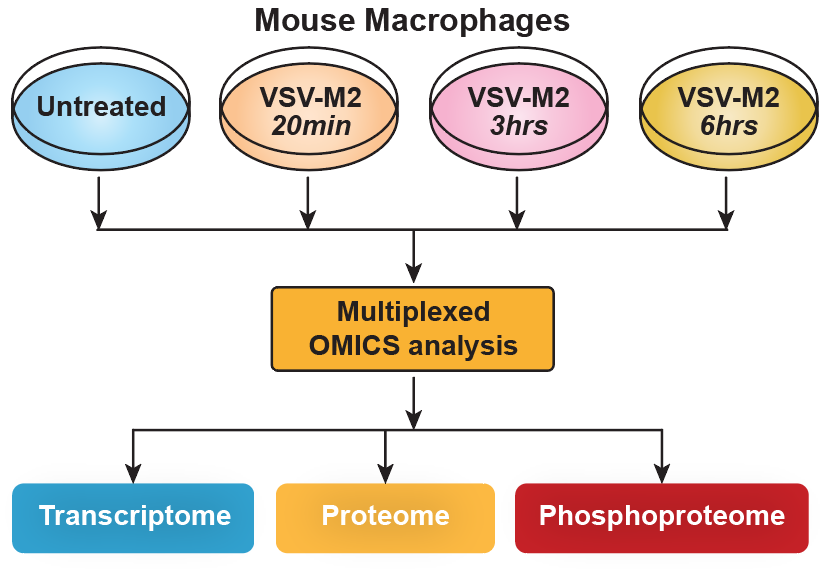
We performed a temporal genome-wide transcriptomics, proteomics and phosphoproteomics analysis of the cellular response of mouse macrophages to Vesicular Stomatitis Virus (VSV) infection. This was followed by integrative bioinformatics analyses to get a global overview of the cellular response (Figure 2).
In practice we sampled the macrophage response at times 20 minutes, 3 hours and 6 hours after infection.
We discovered that immune cells have multiple regulatory options during antiviral response. A novel phosphorylation site as well as four other genes were functionally validated for their role in type-I interferon activation, NFkB activation and VSV life cycle.
The vast and complex molecular changes measured could be decomposed in a limited number of clusters within each category (transcripts, proteins, protein phosphorylation), each with its own kinetic parameters and characteristic pathways and processes, suggesting multiple regulatory options and a specific process logic within the overall sensing and homeostatic program.
Phosphorylation is crucial for tailor-made defence
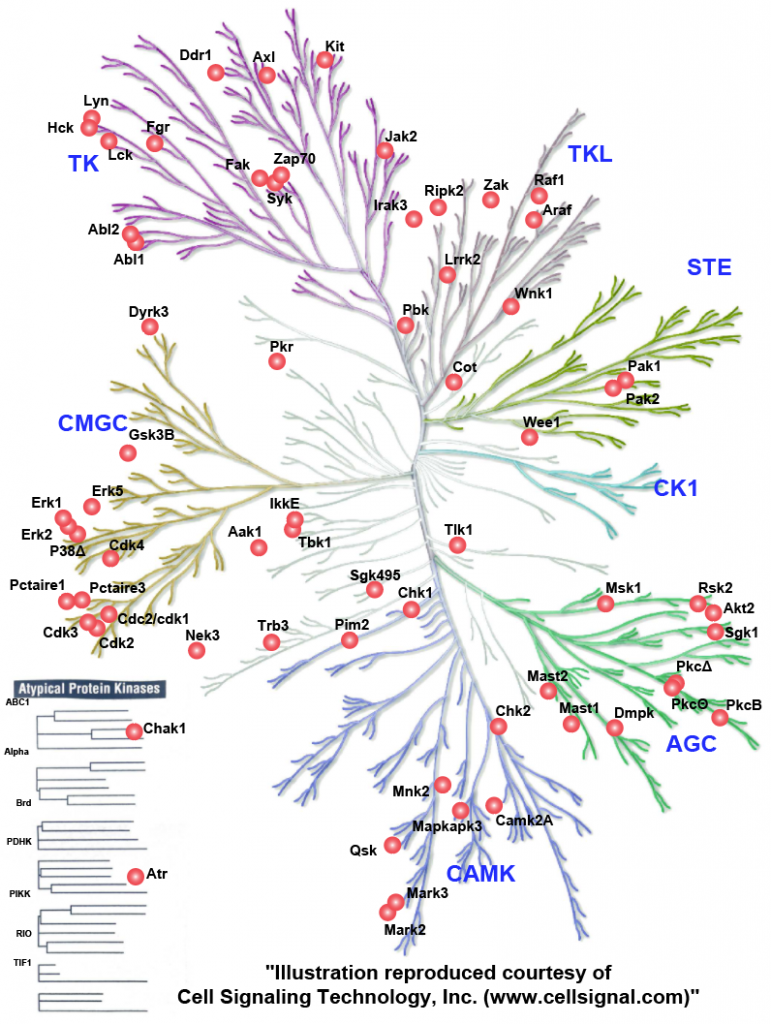
Overall, the data highlighted a predominant executive function to phosphorylation, likely evolved due to the requirement of a fast response to pathogens. Functional validation of a novel phosphorylation site S328-S330 on the innate immunity adaptor MAVS, identified its essential role in activation of type-I interferon and NFkB response. Further, we evaluated the kinase-substrate relationships (Figure 4) and identified RAF1, and to a smaller degree, ARAF to be suppressing VSV replication and needed for NFκB activation, and AKT2 to be favouring VSV replication. Integrative analysis of the omics data showed coregulation of membrane transporters including SLC7A11 which we validated as a host factor in the VSV life cycle.
Open access to the data
The results of the study are published in ” A time-resolved molecular map of the macrophage response to VSV infection in Nature – Systems Biology and Applications . The dataset is presented, and freely available, on the website www.infectome-map.org and represents a large and unique starting platform for further systems-level as well as targeted mechanistic investigations on the functional organization of the response of macrophages to viral infection.